
Robert Haas
Project OwnerProject Manager (Proposal), Research Lead (Review of existing methods), Software Designer & Developer (Package implementation), Technical Writer (Documentation and Report)
DEEP Connects Bold Ideas to Real World Change and build a better future together.
Coming Soon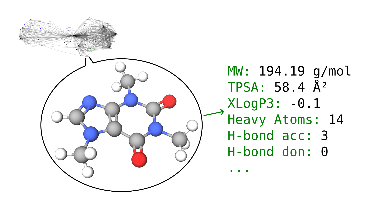
Milestone Release 1 |
$10,000 USD | Pending | TBD |
Milestone Release 2 |
$10,000 USD | Pending | TBD |
Milestone Release 3 |
$23,000 USD | Pending | TBD |
Milestone Release 4 |
$15,000 USD | Pending | TBD |
Milestone Release 5 |
$12,000 USD | Pending | TBD |
Biomedical knowledge graphs (BMKG) contain chemical compounds such as drugs, toxins, metabolites, cofactors or signaling molecules. These entities and some of their relations can be richly augmented with qualitative & quantitative properties by methods from cheminformatics, computer-aided drug design (CADD) and related fields. This enables numerical queries and many analyses such as filtering, clustering, embedding, similarity/outlier detection, QSAR modeling, ML, etc. The aim of this project is to make existing but scattered methods available in a Python package with a unified functional API, expose it to OpenCog Hyperon, and apply it in a PoC study to annotate and analyze Hetionet in MORK.
This RFP seeks the development of advanced tools and techniques for interfacing with, refining, and evaluating knowledge graphs that support reasoning in AGI systems. Projects may target any part of the graph lifecycle — from extraction to refinement to benchmarking — and should optionally support symbolic reasoning within the OpenCog Hyperon framework, including compatibility with the MeTTa language and MORK knowledge graph. Bids are expected to range from $10,000 - $200,000.
New reviews and ratings are disabled for Awarded Projects
Check back later by refreshing the page.
1) Set up and sign the contract. 2) Extend the preliminary research done as prepartion for this proposal. This means a broad literature and code review of existing functionality in cheminformatics and computer-aided drug discovery today will be performed in order to find out what implementations are actively maintained, backed by publications, deliver reliable results, and can be used in combination in a shared conda environment.
The results of the broad literature and code review are provided in form of a GitHub repository rather than a PDF report so that other researchers can find and extend it in an easy way. If determined suitable, it may adhere to the style of an "awesome list" repository to make it easily findable and recognizable.
$10,000 USD
Decide which 5 to 7 external projects are going to be covered and what structure the unified API is going to take. The precise number of projects will depend on how much functionality each of them provides and how complex it is to abstract it into a functional interface that maximizes mutual compatibility. From the current point of view, good candidates seem to be OpenBabel, RDKit, Indigo and PaDeL-Descriptor, which cover a wide range of functionality, e.g. format conversions, descriptor and fingerprint calculations, 3D structure generation, 2D and 3D visualization, tautomer enumeration, etc. Reasonable additions from the perspective of broad methodological coverage could be a dedicated 3D conformer generator like Balloon, a molecular docking program like AutoDock Vina, and perhaps a quantum chemical toolkit like Psi4 for slower but more accurate geometry and electronic structure prediction that could be used as basis for molecular dynamics simulation.
The results of the project selection and API design will be a Python package with a scaffold for the covered toolkits and a few sample functions already implemented to ensure the outline works as intended.
$10,000 USD
Fully implement the Python package that provides a unified API. The aim is to cover a large portion of the methods provided in the chosen external projects, though some very specific methods may not be of broad interest and therefore omitted.
Completed Python package, including a test suite with high coverage of the codebase, and everything required for distributing it as easily installable package on a suitable repository.
$23,000 USD
Apply the Python package on a proof-of-concept case study. A reasonably sized biomedical knowledge graph will be ported to MeTTa and then augmented with various functions provided in the package, either by manual or automatic registering them as grounded atoms in OpenCog Hyperon. Ideally MORK will be used as backend if the project is mature enough at this point. The goal is not only to annotate the BMKG but also to perform interesting queries and analyses on it, e.g. basic filtering up to embedding and QSAR modeling or supervised ML.
Proof-of-concept case study that applies the package on a BMKG such as Hetionet and performs some downstream analyses.
$15,000 USD
Generate a technical documentation website for the Python package and a summary PDF report for the entire project.
Code documentation so that external developers can use it without further help. PDF report so the project and its results can be understood by anyone interested.
$12,000 USD
Please create account or login to post comments.
Reviews & Ratings
New reviews and ratings are disabled for Awarded Projects
Check back later by refreshing the page.
© 2026 DEEP Funding
Sort by